In 1887, Julius Petri invented a couple of glass dishes, designed to grow bacteria in a reproducible, consistent environment. The Petri dish, as it came to be known, birthed the scientific practice of agar cultures, allowing scientists to study bacteria and viruses. The field of microbiology was able to flourish with this handy new tool. The Petri dish, along with advancements in our understanding of microbiology, later developed into the modern field of microbial testing, allowing scientists to understand and measure microbial colonies to detect harmful pathogens in our food and water, like E. coli and Salmonella, for example.
The global food supply chain moves much faster today than it did in the late 19th century. According to Milan Patel, CEO of PathogenDx, this calls for something a little quicker. “Traditional microbial testing is tedious and lengthy,” says Patel. “We need 21st century pathogen detection solutions.”
Milan Patel first joined the parent company of PathogenDx back in 2012, when they were more focused on clinical diagnostics. “The company was predominantly built on grant funding [a $12 million grant from the National Institute of Health] and focused on a niche market that was very specialized and small in terms of market size and opportunity,” says Patel. “I realized that the technology had a much greater opportunity in a larger market.”

Photo: Michael Chansley
He thought that other markets could benefit from that technology greatly, so the parent company licensed the technology and that is how PathogenDx was formed. Him and his team wanted to bring the product to market without having to obtain FDA regulatory approval, so they looked to the cannabis market. “What we realized was we were solving a ‘massive’ bottleneck issue where the microbial test was the ‘longest test’ out of all the tests required in that industry, taking 3-6 days,” says Patel. “We ultimately realized that this challenge was endemic in every market – food, agriculture, water, etc. – and that the world was using a 140-year-old solution in the form of petri dish testing for microbial organisms to address challenges of industries and markets demanding faster turnaround of results, better accuracy, and lower cost- and that is the technology PathogenDx has invented and developed.”
While originally a spinoff technology designed for clinical diagnostics, they deployed the technology in cannabis testing labs early on. The purpose was to simplify the process of testing in an easy approach, with an ultra-low cost and higher throughput. Their technology delivers microbial results in less than 6 hours compared to 24-36 hours for next best option.

Out of all the tests performed in a licensed cannabis testing laboratory, microbial tests are the longest, sometimes taking up to a few days. “Other tests in the laboratory can usually be done in 2-4 hours, so growers would never get their microbial testing results on time,” says Patel. “We developed this technology that gets results in 6 hours. The FDA has never seen something like this. It is a very disruptive technology.”
When it comes to microbial contamination, timing is everything. “By the time Petri dish results are in, the supply chain is already in motion and products are moving downstream to distributors and retailers,” Patel says. “With a 6-hour turnaround time, we can identify where exactly in the supply chain contaminant is occurring and spreading.”
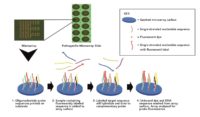
The technology is easy to use for a lab technician, which allows for a standard process on one platform that is accurate, consistent and reproduceable. The technology can deliver results with essentially just 12 steps:
- Take 1 gram of cannabis flower or non-flower sample. Or take environmental swab
- Drop sample in solution. Swab should already be in solution
- Vortex
- Transfer 1ml of solution into 1.5ml tube
- Conduct two 3-minute centrifugation steps to separate leaf material, free-floating DNA and create a small pellet with live cells
- Conduct cell lysis by adding digestion buffer to sample on heat blocks for 1 hour
- Conduct Loci enhancement PCR of sample for 1 hour
- Conduct Labelling PCR which essentially attaches a fluorescent tag on the analyte DNA for 1 hour
- Pipette into the Multiplex microarray well where hybridization of sample to probes for 30 minutes
- Conduct wash cycle for 15 minutes
- Dry and image the slide in imager
- The imager will create a TIFF file where software will analyze and deliver results and a report
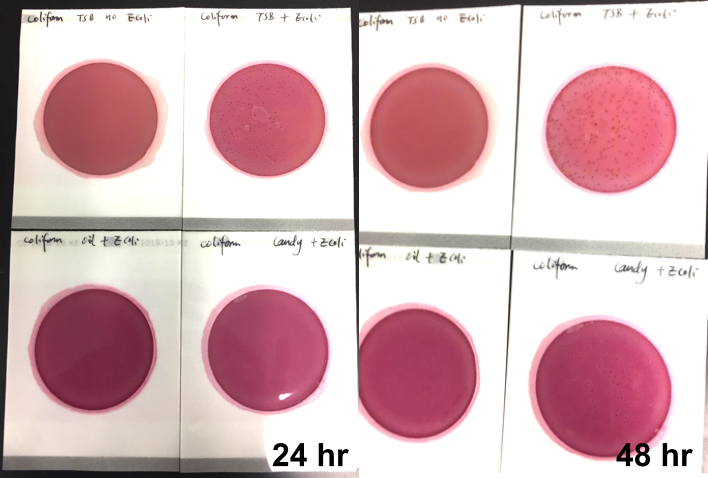
Their DetectX product can test for a number of pathogens in parallel in the same sample at the same time down to 1 colony forming unit (CFU) per gram. For bacteria, the bacterial kit can detect E. coli, E. coli/Shigella spp., Salmonella enterica, Listeria and Staph aureus, Stec 1 and Stec 2 E.coli. For yeast and mold, the fungal kit can test for Aspergillus flavus, Aspergillus fumigatus, Aspergillus niger and Aspergillus terreus.
Their QuantX is the world’s first and only multiplex quantification microarray product that can quantify the microbial contamination load for key organisms such as total aerobic bacteria, total yeast & mold, bile tolerant gram negative, total coliform and total Enterobacteriaceae over a dynamic range from 100 CFU/mL up to 1,000,000 CFU/mL.
Not all of the PathogenDx technology is designed for just microbial testing of cannabis or food products. Their EnviroX technology is designed to help growers, processors or producers across any industry identify areas of microbial contamination, being used as a tool for quality assurance and hazard analysis. They conducted industry-wide surveys of the pathogens that are creating problems for cultivators and came up with a list of more than 50 bacterial and fungal pathogens that the EnviroX assay can test for to help growers identify contamination hotspots in their facilities.
 Using the EnviroX assay, growers can swab surfaces like vents, fans, racks, workbenches and other potential areas of contamination where plants come in contact. This helps growers identify potential areas of contamination and remediate those locations. Patel says the tool could help growers employ more efficient standard operating procedures with sanitation and sterilization, reducing the facility’s incidence of pathogens winding up on crops, as well as reduction in use of pesticides and fungicides on the product.
Using the EnviroX assay, growers can swab surfaces like vents, fans, racks, workbenches and other potential areas of contamination where plants come in contact. This helps growers identify potential areas of contamination and remediate those locations. Patel says the tool could help growers employ more efficient standard operating procedures with sanitation and sterilization, reducing the facility’s incidence of pathogens winding up on crops, as well as reduction in use of pesticides and fungicides on the product.
Deploying this technology in the cannabis industry allowed Milan Patel and the PathogenDx team to bring something new to the world of microbial testing. Their products are now in more than 90 laboratories throughout the country. The success of this technology provides another shining example of how the cannabis market produces innovative and disruptive ideas that have a major impact on the world, far beyond cannabis itself.