Microbial contamination on cannabis products represents one of the most significant threats to cannabis consumers, particularly immunocompromised patients who are at risk of developing harmful and potentially fatal infections.
As a result, regulatory bodies in the United States and Canada mandate testing cannabis products for certain microbes. The two most popular methods for microbial safety testing in the cannabis industry are culture-based testing and quantitative polymerase chain reaction (qPCR).
When considering patient safety, labs should choose a method that provides an accurate account of what is living on the sample and can specifically target the most harmful microbes, regardless of the matrix.
1. The Method’s Results Must Accurately Reflect the Microbial Population on the Sample
The main objective of any microbial safety test is to give the operator an indication of the microbial population present on the sample.

Culture-based methods measure contamination by observing how many organisms grow in a given medium. However, not all microbial organisms grow at the same rate. In some cases, certain organisms will out-compete others and as a result, the population in a post-culture environment is radically different than what was on the original sample.
One study analyzed fifteen medicinal cannabis samples using two commercially available culture-based methods. To enumerate and differentiate bacteria and fungi present before and after growth on culture-based media, all samples were further subjected to next-generation sequencing (NGS) and metagenomic analyses (MA). Figure 1 illustrates MA data collected directly from plant material before and after culture on 3M petrifilm and culture-based platforms.
The results demonstrate substantial shifts in bacterial and fungal growth after culturing on the 3M petrifilm and culture-based platforms. Thus, the final composition of microbes after culturing is markedly different from the starting sample. Most concerning is the frequent identification of bacterial species in systems designed for the exclusive quantification of yeast and mold, as quantified by elevated total aerobic count (TAC) Cq values after culture in the total yeast and mold (TYM) medium. The presence of bacterial colonies on TYM growth plates or cartridges may falsely increase the rejection rate of cannabis samples for fungal contamination. These observations call into question the specificity claims of these platforms.
The Live Dead Problem

One of the common objections to using qPCR for microbial safety testing is the fact that the method does not distinguish between live and dead DNA. PCR primers and probes will amplify any DNA in the sample that matches the target sequence, regardless of viability. Critics claim that this can lead to false positives because DNA from non-viable organisms can inflate results. This is often called the Live-Dead problem. However, scientists have developed multiple solutions to this problem. Most recently, Medicinal Genomics developed the Grim Reefer Free DNA Removal Kit, which eliminates free DNA contained in a sample by simply adding an enzyme and buffer and incubating for 10 minutes. The enzyme is instantaneously inactivated when lysis buffer is added, which prevents the Grim Reefer Enzyme from eliminating DNA when the viable cells are lysed (see Figure 2).
2. Method Must Be Able to Detect Specific Harmful Species
Toxic Aspergillus spp., which is responsible for at least one confirmed death of a cannabis patient, grows poorly in culture mediums and is severely underreported by current culture-based platforms. And even when Aspergillus does grow in culture, there is a certain non-pathogenic Aspergillus species that look remarkably similar to their pathogenic cousins, making it difficult to speciate using visual identification alone.
Conversely, qPCR assays, such as the PathoSEEK, are designed to target DNA sequences that are unique to pathogenic Aspergillus species, and they can be run using standard qPCR instruments such as the Agilent AriaMx. The primers are so specific that a single DNA base difference in the sequence can determine whether binding occurs. This specificity reduces the frequency of false positives in pathogen detection, a frequent problem with culture-based cannabis testing methods.
Additionally, Medicinal Genomics has developed a multiplex assay that can detect the four pathogenic species of Aspergillus (A. flavus, A. fumigatus, A. niger, and A. terreus) in a single reaction.
3. The Method Must Work on Multiple Matrices

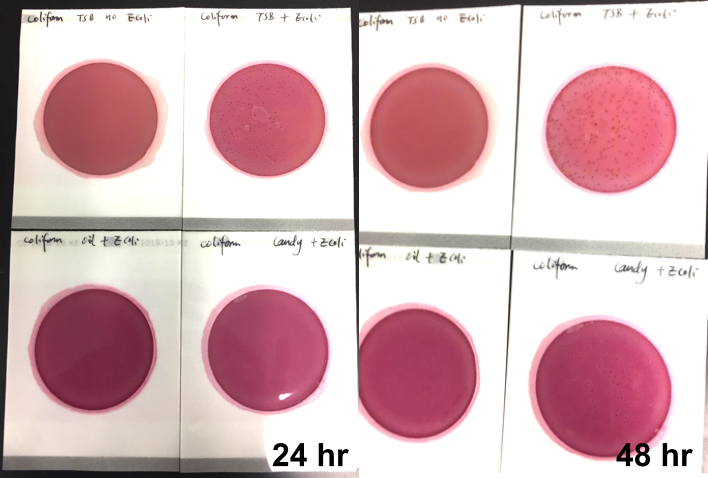
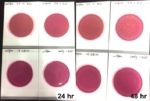
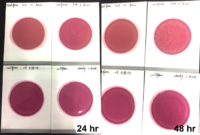
Marijuana infused products (MIPs) are a very diverse class of matrices that behave very differently than cannabis flowers. Gummy bears, chocolates, oils and tinctures all present different challenges to culture-based techniques as the sugars and carbohydrates can radically alter the carbon sources available for growth. To assess the impact of MIPs on colony-forming units per gram of sample (CFU/g) enumeration, The Medicinal Genomics team spiked a known amount of live E. coli into three different environments: tryptic soy broth (TSB), hemp oil and hard candy. The team then homogenized the samples, pipetted amounts from each onto 3M™ Petrifilm E. coli / Coliform Count (EC) Plates, and incubated for 96 hours. The team also placed TSB without any E. coli onto a petrifilm to serve as a control. Figures 3 and 4 show the results in 24-hour intervals.

This implies the MIPs are interfering with the reporter assay on the films or that the MIPs are antiseptic in nature.
Many MIPs use citric acid as a flavoring ingredient which may interfere with 3M reporter chemistry. In contrast, the qPCR signal from the Agilent AriaMx was constant, implying there is microbial contamination present on the films, but the colony formation or reporting is inhibited.


This is not an issue with DNA-based methods, so long as the DNA extraction method has been validated on these matrices. For example, the SenSATIVAx DNA extraction method is efficient in different matrices, DNA was spiked into various MIPs as shown in Table 1, and at different numbers of DNA copies into chocolate (Table 2). The SenSATIVAx DNA extraction kit successfully captures the varying levels of DNA, and the PathoSEEK detection assay can successfully detect that range of DNA. Table 3 demonstrates that SenSATIVAx DNA extraction can successfully lyse the cells of the microbes that may be present on cannabis for a variety of organisms spiked onto cannabis flower samples.