When people have to make important decisions, we often consult a third party to increase our knowledge and confidence in a product. For instance, when choosing a car, an individual may weigh heavily on safety ratings and other awards from organizations such as Consumer Reports. These awards are often boasted and a heavy focus in car commercials because it tells the consumer that a third party has deemed their car valuable to own. For more than 100 years, the Association of Official Agricultural Chemists (AOAC® International) has operated in this exact manner, and has set the bar and guidelines for testing in the cannabis industry through its special program called the Cannabis Analytical Science Program, also known as CASP.
The CASP program is designed to develop standards and validation guidance to evaluate testing methods, as well as the methods’ ability to detect the target organism or compound on the cannabis matrix. With the addition of new states permitting the legal sale of both medical and adult use cannabis and no federal governing body overseeing testing regulations, the value of AOAC cannot be understated, as these guidelines allow cannabis testing laboratories to have their own third-party reference to look to when choosing a compliant testing method to implement in their laboratory.
![]() AOAC was founded in 1884 by the US government as the standard setting body in the country and, in 1991, became an independent association known as AOAC International, with a goal of building a reputation as an international, consensus-based standard-setting body and a conformity assessment organization in analytical sciences. As an independent third-party resource, AOAC has the Performance Tested Methods’ (PTM) and Official Methods of AnalysisSM (OMA) programs for certification of analytical testing methods in both biology and chemistry.
AOAC was founded in 1884 by the US government as the standard setting body in the country and, in 1991, became an independent association known as AOAC International, with a goal of building a reputation as an international, consensus-based standard-setting body and a conformity assessment organization in analytical sciences. As an independent third-party resource, AOAC has the Performance Tested Methods’ (PTM) and Official Methods of AnalysisSM (OMA) programs for certification of analytical testing methods in both biology and chemistry.
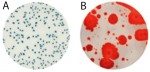
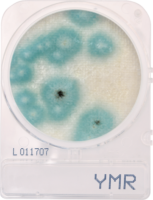
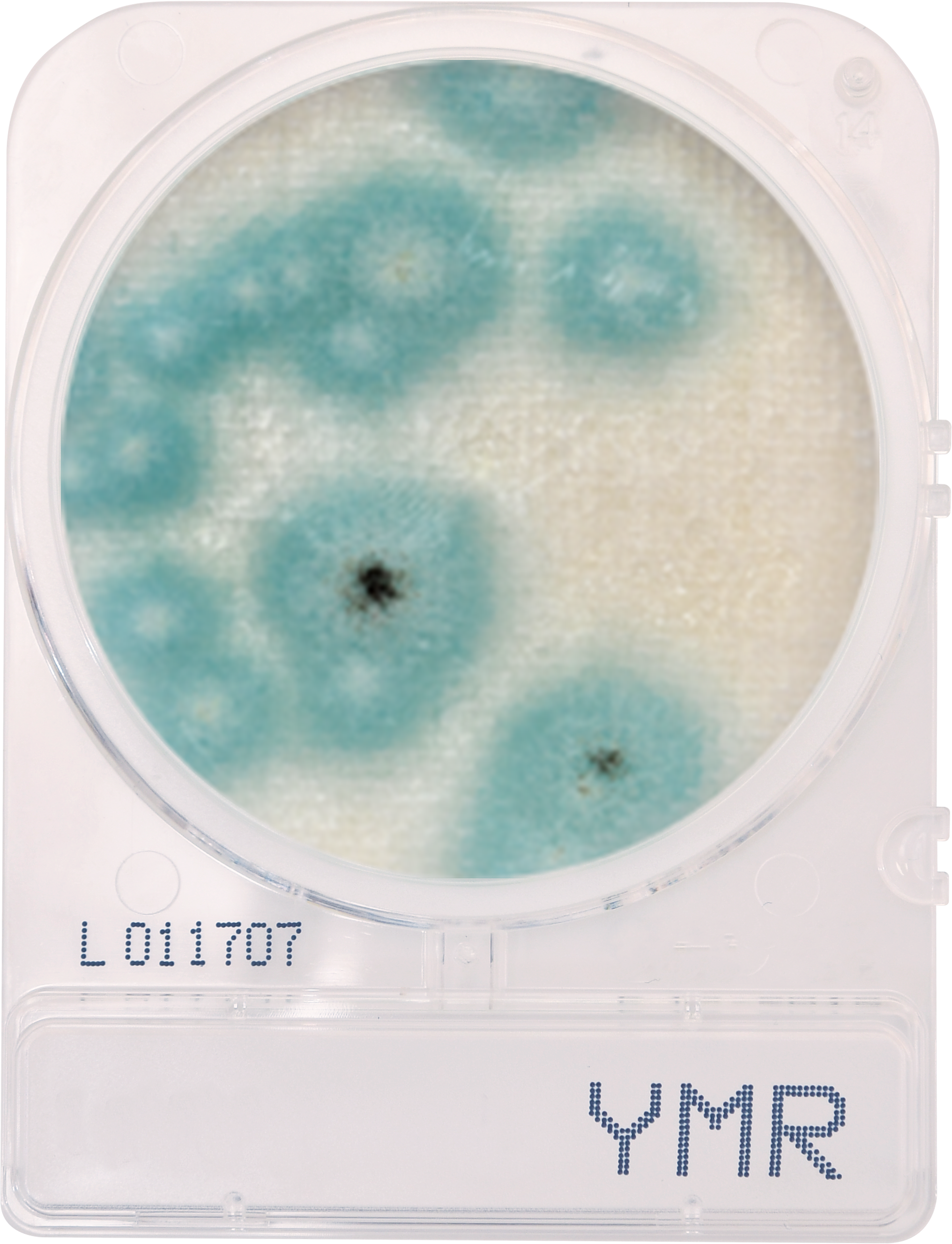
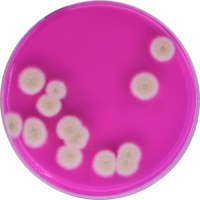
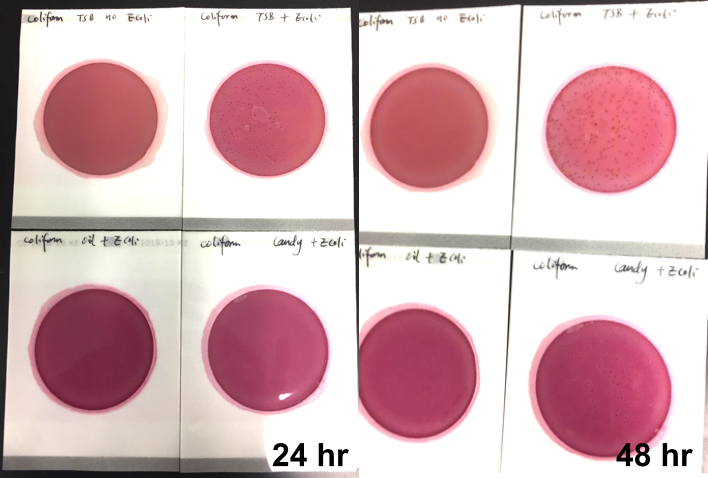
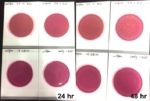
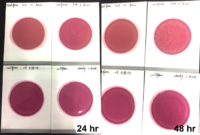
If analytical methods, including proprietary test kits, are deemed acceptable, AOAC provides approved certification, their seal of approval that the method works as designed. Though multiple factors are considered to determine if AOAC approval is given; accuracy and precision of the method are among the most important. For example, when validating a cannabis method for microbiology, AOAC will contract an independent testing facility to conduct a series of tests with known spiked samples to measure the recovery limit of the target microorganism. This allows the organization to determine if the method is sensitive enough to be named an AOAC-approved method through either the PTM or OMA conformity programs. Another way of ensuring the validity of results is by conducting an inclusivity and exclusivity study on a method. In this type of experiment, target organisms are tested while also spiking with non-target organisms to see if there will be a high rate of false positives.
 In cannabis, discussions have grown surrounding testing of four strains of Aspergillus, which are A. terreus, A. flavus, A. fumigatus and A. niger. By spiking cannabis with one of the four Aspergillus strains and on a separate sample with a non-target Aspergillus strain such as A. clavatus, it ensures that only the target strains are being recognized and recorded on the method being tested.
In cannabis, discussions have grown surrounding testing of four strains of Aspergillus, which are A. terreus, A. flavus, A. fumigatus and A. niger. By spiking cannabis with one of the four Aspergillus strains and on a separate sample with a non-target Aspergillus strain such as A. clavatus, it ensures that only the target strains are being recognized and recorded on the method being tested.
This methodology limits the likelihood of unconfirmed positives occurring, ensuring the validity of the results. Of course, when a method is undergoing an actual AOAC evaluation for approval, the testing requirements for both the sensitivity and inclusivity/exclusivity experiments are much more thorough than the explanation above.
Regardless of which AOAC-approved method you select, you can feel confident that most of the “heavy-lifting” is done and that the method is accurate and precise enough to implement in a cannabis testing facility. In turn, the cannabis testing laboratory then only needs to complete their own internal method verification to ensure the method works with their processes, people, environment and product, but on a much smaller scale and aligns with state regulations.
 On a consumer safety level, AOAC-approved methods are designed to keep cannabis consumers safe. Whether they are an adult using cannabis or medicinal cannabis patient, the product that is being sold should be held to the highest safety standards. By having a laboratory that is utilizing an independently approved AOAC method, an additional layer of confidence is achieved that the product being consumed is safe. This ultimately limits the number of costly recalls from dispensaries and minimizes risk to consumers. At the end of the day, cannabis testing laboratories want to keep the public safe and it is our job to do so. This means implementing these independently approved methods from agencies such as AOAC at various touch points in the seed to sale cycle to ensure the data is validated and reliable.
On a consumer safety level, AOAC-approved methods are designed to keep cannabis consumers safe. Whether they are an adult using cannabis or medicinal cannabis patient, the product that is being sold should be held to the highest safety standards. By having a laboratory that is utilizing an independently approved AOAC method, an additional layer of confidence is achieved that the product being consumed is safe. This ultimately limits the number of costly recalls from dispensaries and minimizes risk to consumers. At the end of the day, cannabis testing laboratories want to keep the public safe and it is our job to do so. This means implementing these independently approved methods from agencies such as AOAC at various touch points in the seed to sale cycle to ensure the data is validated and reliable.
Overall, just as it is equally important to get a non-biased and reputable third-party approach to your automobile search, a scientist that is responsible for choosing methods in their cannabis compliance laboratory should also consider these third-party approvals. As a scientist, the goal every day is to report accurate data to help the client and the consumer equally. The cannabis compliance laboratories are the last line of defense in preventing harmful or contaminated products from getting into the marketplace and any extra assurance we have with our testing methodology is always encouraged. Ultimately, AOAC’s work is important and their standard of quality and safety is a must-have in the cannabis laboratory.