Inexpensive in vitro Methods to Evaluate the Impact of Cannabinoid-containing Products on Sentinel Lactobacillus spp.
S. Lewin 1, A. Hilyard2, H. Piscatelli1, A. Hangman1, D. Petrik1, P. Miles2, and C. Orser2
1MatMaCorp Inc, Lincoln NE; 2Apothercare LLC, Boston MA
Abstract
The public has readily embraced cannabidiol (CBD) in countless unregulated products that benefit from commercial promotion without FDA oversight, who recently concluded: “that a new regulatory pathway for CBD is needed that balances individuals’ desire for access to CBD products w/ the regulatory oversight is needed to manage risks.”1 The reported antimicrobial properties of CBD combined with the recent proliferation of cannabinoid-containing products marketed to women for intimate care led us to explore the impact on the sentinel lactobacilli species associated with a healthy reproductive tract. Except for lubricants and tampons, the FDA regulates intimate care products as cosmetics. Even non-cannabis serums, washes, and suppositories are not required to be tested for their effect on the reproductive microbiota. We aimed to investigate the utility of easy-to-use, inexpensive in vitro assays for testing exogenous cannabis products on reproductive microbiota. In vitro assays can provide important evidence-based data to inform both manufacturers choosing both an active cannabinoid ingredient source as well as excipient chemicals and consumers in the absence of safety or quality data. In simple, straightforward exposure studies, we examined the antimicrobial activity of CBD and cannabigerol (CBG) on the most dominant vaginal lactobacilli species, L. crispatus, associated with good health.
Introduction
The testing of readily available products containing cannabinoids, predominately CBD following the widespread legalization of hemp by the 2018 US Farm Bill, is not required beyond ensuring THC content is below 0.3%. Therefore, basic information on safety, quality, antimicrobial activity, bioavailability, and dosing is unavailable and undocumented. The situation is further complicated by the complex chemoprofiles of cannabis extracts based on the cultivar, the extraction methods and subsequent cleanup, and other chemical excipients in the formulation. The FDA has finalized guidance on quality considerations for clinical research for the development of cannabis and cannabis-containing drugs intended for human use.
One approach to backfilling non-existent safety and quality data for cannabinoid active ingredients and those products made from them is to apply or devise assays that can provide relevant toxicity data in an in vitrosystem. Farha et al. (2020) reported that seven cannabinoids are potent antibiotics, including CBD and synthetic CBG. CBG inhibited the growth of gram-positive bacteria, including methicillin-resistant Staphylococcus aureus (MRSA), but not gram-negative bacteria unless their outer membrane was permeabilized (Farha et al. 2020). In addition, several volatile terpenes, the main constituents of essential oils extracted from Cannabis sativa L., also have potent antibiotic activity against gram-positive bacteria (Iseppi et al. 2019). We have previously written about the risks associated with disrupting the healthy microbiome of gram-positive vaginal bacterial species leading to dysbiosis (Orser 2022) and its further health complications.
Several successful approaches to assessing the toxicity of CBD have already been reported including human cell culture work by Torres et al. (2022) who showed that pure CBD has a repeatable impact on cell viability, but that hemp-derived finished CBD products had variable impact. Cultured human cell viability experiments demonstrated similar potencies across three different hemp-derived CBD products in the microgram per milliliter [mg/mL] range with increased viability at lower doses [2-4 mg/mL] and decreasing cell viability above 6 mg/mL (Torres et al. 2022). In the same study, the authors demonstrated that the presence of terpenes, specifically b-caryophyllene, in hemp extraction matrices also impacted cell viability.
Neswell, a cannabis therapeutics company in Israel, demonstrated through the application of their in vitroneutrophil cell line that cannabis extracts have inherent immune response biodiversity, suggesting that the choice of a cannabis source should be based on its function rather than on its chemoprofile (https://www.neswell.net). Inflammatory cytokine levels in inflamed peripheral blood mononuclear cells (PB_MC) showed a 10-fold difference across hemp extract products containing unidentified terpenes in suppressing the inflammatory cytokine, TNFa (Torres et al. 2022). The influence of CBD concentration on inflammatory cytokine production was previously reported by Vuolo et al. (2015) and Jiang et al. (2022).
Materials & Methods
Chemicals and Products Tested
THC-free, 99% pure CBD and CBG isolates were purchased from Open Book extracts in North Carolina (openbookextracts.com). All other chemicals including erythromycin (EM), and growth media were obtained from Sigma-Aldrich (St. Louis MO). Specific reagents in the qPCR kits were assembled in-house at MatMaCorp Inc. (Lincoln NE).
Monitoring Cell Viability: OD600nm and plating
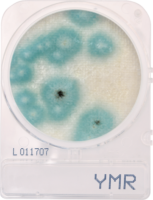
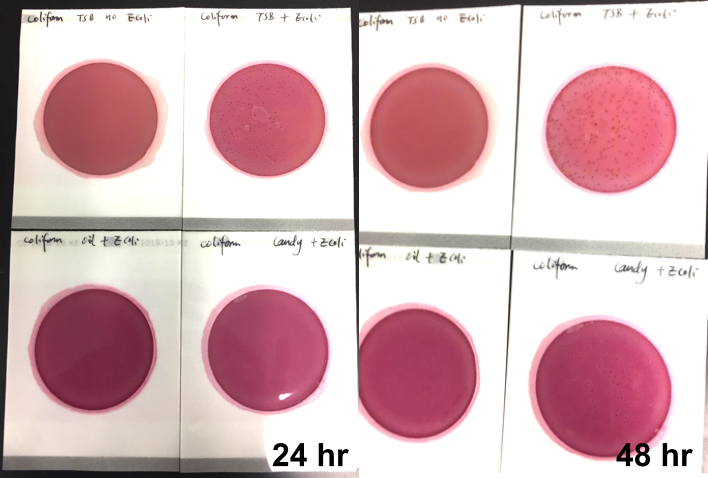
Individual frozen glycerin stocks of L. crispatus HM103 from BEI Resources Repository served as inoculum to streak on a sterile MRS agar plate and incubated anaerobically at 370C for 24-48 h until individual colony growth was observed. Single colonies were used to inoculate MRS broth and incubated for 24-48 h at 370C which served as the inoculum for exposure to test products. Exposed cultures and all control cultures were incubated at 370C for 48 h with OD600 readings taken at time zero, +24 h, and +48 h using disposable cuvettes in a standard spectrophotometer. The products were also plated onto MRS agar plates to evaluate inherent contaminants that could affect turbidity values.
Molecular Analysis by qPCR
DNA isolation from bacterial cultures was done using the MatMaCorp (Lincoln, NE) StickE Tissue DNA Isolation kit modified for bacteria as per manufacturer instructions. Briefly, a lysis buffer is applied to the sample followed by a heating step, and a binding buffer is added, thus allowing DNA from the solution to bind to the matrix of the StickE column. The column was washed prior to eluting the purified DNA. Per manufacturer instructions, 10 µL of isolated DNA was used as a template for genetic analysis in a Lacto-TM assay (MatMaCorp). The assay is a customized TaqMan-based detection assay that is conducted using a four-channel fluorescence detection platform, the Solas 8 (MatMaCorp). The assay was designed to detect the unique 16S-rRNA DNA sequence for L. crispatus. Briefly, the assay is a probe-based method that begins with hybridizing the custom-designed probes with their desired nucleic acid target found in the sample. Once hybridized, detection takes place from the fluorescently labeled primer. The target has been assigned a channelon the Solas 8 and is detected independently.
Calling the Results
The calling algorithm uses first-order kinetics reaction properties (inflection point detection) in combinationwith a measure of the closeness of the signals associated with a specific target. Various indicators are tracked during the reactions to perform an on-the-fly analysis. The analysis is then consolidated by a measure of the similarity between the fluorescence signals at the end of the run. Aggregating values from the similarity measure, the end gain and the inflection point detection allow the Solas 8 software to make the call at the end of the run without having to compare a results library of known sample targets.

Results
Exposure of L. crispatus
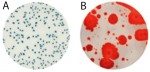
Anaerobically grown cultures of L. crispatus were exposed to either CBD isolate or CBG isolate at each of two concentrations [5 mg/mL] and [10 mg/mL] with all appropriate controls. All treatment groups were evaluated by qPCR, turbidity at OD600, and plate counts.
Molecular Analysis via qPCR
These data show the specificity of the Solas8 testing for evaluating these products, as a molecular-level screening is not influenced by test product solubility, opacity, or non-specific contamination present in some of the tested products that can interfere with optical density measurements.
Growth Monitoring
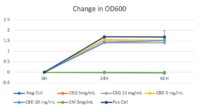
Turbidity monitoring, albeit non-specific, confirmed the species-specific qPCR findings, that is no inhibition for the two cannabinoid isolates evaluated (Fig. 2).
Conclusions
In this limited in vitro study using a sentinel lactobacilli response, we have shown that 99% pure CBD and CBG isolates were not inhibitory at the two doses evaluated by complementary observations following turbidity, plating, and by qPCR. Limitations in this study prevent definitive conclusions regarding what individual or combination of cannabinoids or other cannabis secondary metabolites are inhibitory in vivo to dominant lactobacilli species in the reproductive tract. These limitations include commercial product testing without knowledge of excipients or impact on the bioavailability of any active cannabinoid ingredients. In addition, dose-response curves were not generated and exposure under micro-aerobic conditions was not carried out.
Cannabidiol’s potential as an antimicrobial agent may be limited by its extremely low solubility in water and a propensity to stick to spurious proteins limiting systemic distribution in the body as a therapeutic. Interpreting microbiome study findings to human health outcomes will require multi-disciplinary corresponding clinical data findings of disease diagnosis, processes, and treatment within populations. Nonetheless, this nascent translational research opportunity is vast with the promise of benefiting patient outcomes (Wensel et al. 2022).
Health Canada released a scientific review report on products containing cannabis, specifically containing 98% or greater CBD and less than 1% of THC (Health Canada 2022) while the FDA just concluded that there are no existing guidelines applicable for recommending safety and quality guidelines to manage risk for CBD products (U.S. FDA 2023). The Health Canada committee unanimously agreed that short-term use of CBD is safe at 20 mg per day up to a maximum dose of 200 mg per day and that packaging should include both dosing instructions and potential side effects. The Committee did not address the antimicrobial potential of CBD or CBG formulations or specifically vulvar or vaginally administered cannabinoids. There is clearly more basic physiological research needed on the impact of self-administration of CBD preparations based on the route of exposure.
References
Farha MA, El-Halfawy LM, Gale RT, MacNair CR, Carfrae LA, Zhang X, Jentsch NG, Magolan J, Brown ED (2020) Uncovering the hidden antibiotic potential of cannabis. ACS Infect Dis 6:338-346.
Health Canada (2022). https://www.canada.ca/content/dam/hc-sc/documents/corporate/about-health-canada/public-engagement/external-advisory-bodies/health-products-containing-cannabis/report-cannabidiol-eng.pdf
Hopkins AL (2008) Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol 4(11):682-90.
Iseppi R, Brighenti V, Licata M, Lambertini A, Sabia C, Messi P, Pellati F, Benvenuti S (2019) Chemical characterization and evaluation of the antibacterial activity of essential oils from fibre-type Cannabis sativa L. (Hemp) Molecules 24:2302; doi:10.3390/molecules24122302.
Jiang Z, Jin S, Fan X, Cao K, Liu Y, Want X, Ma Y, Xiang L (2022) Cannabidiol inhibits inflammation induced by Cutibacterium acnes-derived extracellular vesicles via activation of CB2 receptor in keratinocytes. J Inflammation Res 15:4573-4583.
Orser CS (2022) Prevalence of Cannabinoid-containing Intimate Care Products Exposes Longstanding Unmet Need for Safety Data on Community Microbiota Exposure. https://cannabisindustryjournal.com/feature_article/intimate-care-products-with-cannabinoids-need-more-safety-data/
Torres AR, Caldwell VD, Morris S, Lyon R (2022) Human cells can be used to study cannabinoid dosage and inflammatory cytokine responses. Cannabis Sci & Tech 5(2) 38-45).
U.S. FDA (2023) https://www.fda.gov/news-events/press-announcements/fda-concludes-existing-regulatory-frameworks-foods-and-supplements-are-not-appropriate-cannabidiol
Vuolo F, Petronilho F, Sonai B, Ritter C, Hallak JE, Zuardi AW, Crippa JA, Dal-Pizzol F (2015) Mediators Inflamm 538670
Wensel CR, Salzberg SL, Sears CL (2022) Next-generation sequencing insights to advance clinical investigations of the microbiome. J Clin Invest 132(7):e154944. https://doi.org/10.1172/JCI154944.