Cannabis testing to detect microbial contamination is complicated. It may not be rocket science, but it is life science, which means it’s a moving target, or at least, it should be, as we acquire more and more information about how the world we live in works. We are lucky to be able to carry out that examination in ever increasing detail. For instance, the science of genomics1 was born over 80 years ago, and just twenty years ago, genetics was still a black box. We’ve made tremendous progress since those early days, but we still have a long way to go, to be sure.
Much of that progress is due to our ability to build more accurate tools, a technological ladder, if you will, that raises our awareness, expertise, and knowledge to new levels. When a new process or technology appears, we compare it against accepted practice to create a new paradigm and make the necessary adjustments. But people have to be willing to change. In the cannabis industry, rapid change is a constant, first because that is the nature of a nascent industry, and second because in the absence of some universal and unimpeachable standard, it’s difficult to know who’s right. Especially when the old, reliable reference method (i.e. plating, which is basically growing microorganisms on the surface of a nutritional medium) is deeply flawed in its application to cannabis testing vs. molecular methods (i.e., quantitative polymerase chain reaction, or qPCR for short).

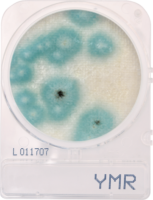
Plating systems have been used faithfully for close to 130 years in the food industry, and has performed reasonably well.2 But cannabis isn’t food and can’t be tested as if it were. In fact, plating methods have a host of major disadvantages that only show up when they’re used to detect cannabis pathogens. They are, in no particular order:
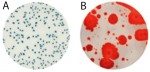
- A single plating system can’t enumerate a group of microorganisms and/or detect specific bacterial and fungal pathogens. This is further complicated by the fact that better than 98% of the microbes in the world do not form colonies.3 And there is no ONE UNIVERSAL bacterial or fungal SELECTIVE agar plate that will allow the growth of all bacteria or all fungal strains. For example, the 5 genus species of fungal strains implicated in powderly mildew DO NOT plate at all.
- Cannabinoids, which can represent 10-30% of a cannabis flower’s weight, have been shown to have antibacterial activity.4 Antibiotics inhibit the growth of bacteria and in some cases kill it altogether. Salmonella species & shiga toxin producing coli (STEC) bacteria, in particular, are very sensitive to antibiotics, which leads to either a false negative result or lower total counts on plates vs. qPCR methods.
- Plating methods cannot detect bacterial and fungal endophytes that live a part or all of their life cycle inside a cannabis plant.5,6 Examples of endophytes are the Aspergillus pathogens (A. flavus, A. fumigatus, A. niger, and A. terreus). Methods to break open the plant cells to access these endophytes to prepare them for plating methods also lyse these microbial cells, thereby killing endophytic cells in the process. That’s why these endophytes will never form colonies, which leads to either false negative results or lower total counts on plates vs. qPCR methods.
- Selective plating media for molds, such as Dichloran Rose-Bengal Chloramphenicol (DRBC) actually reduces mold growth—especially Aspergillus—by as much as 5-fold.This delivers false negative results for this dangerous human pathogen. In other words, although the DRBC medium is typically used to reduce bacteria; it comes at the cost of missing 5-fold more yeast and molds than Potato Dextrose Agar (PDA) + Chloramphenicol or molecular methods. These observations were derived from study results of the AOAC emergency response validation.7
- Finally, we’ve recently identified four bacterial species, which are human pathogens associated with cannabis that do not grow at the plating system incubation temperature typically used.8 They are Aeromonas hydrophila, Pantoea agglomerans, Yersinia enterocolitica, and Rahnella aquatilis. This lowers total counts on plates qPCR methods.
So why is plating still so popular? Better yet, why is it still the recommended method for many state regulators? Beats me. But I can hazard a couple of guesses.
First, research on cannabis has been restricted for the better part of the last 70 years, and it’s impossible to construct a body of scientific knowledge by keeping everyone in the dark. Ten years ago, as one of the first government-employed scientists to study cannabis, I was tapped to start the first cannabis testing lab at the New Jersey Dept. of Health and we had to build it from ground zero. Nobody knew anything about cannabis then.
Second, because of a shortage of cannabis-trained experts, members of many regulatory bodies come from the food industry—where they’ve used plating almost exclusively. So, when it comes time to draft cannabis microbial testing regulations, plating is the default method. After all, it worked for them before and they’re comfortable with recommending it for their state’s cannabis regulations.
Finally, there’s a certain amount of discomfort in not being right. Going into this completely new area—remember, the legal cannabis industry didn’t even exist 10 years ago—we human beings like to have a little certainty to fall back on. The trouble is, falling back on what we did before stifles badly needed progress. This is a case where, if you’re comfortable with your old methods and you’re sure of your results, you’re probably wrong.
So let’s accept the fact that we’re all in this uncharted territory together. We don’t yet know everything about cannabis we need to know, but we do know some things, and we already have some pretty good tools, based on real science, that happen to work really well. Let’s use them to help light our way.
References
- J. Weissenbach. The rise of genomics. Comptes Rendu Biologies, 339 (7-8), 231-239 (2016).
- R. Koch. 1882. Die Aetiologie der Tuberculose. Berliner Klinische Wochenschrift, 19, 221-230 (1882).
- W. Wade. Unculturable bacteria—the uncharacterized organisms that cause oral infections. Journal of the Royal Society of Medicine, 95(2), 91-93 (2002).
- J.A. Karas, L.J.M. Wong, O.K.A. Paulin, A. C. Mazeh, M.H. Hussein, J. Li, and T. Vekov. Antibiotics, 9(7), 406 (2020).
- M. Taghinasab and S. Jabaji, Cannabis microbiome and the role of endophytes in modulating the production of secondary metabolites: an overview. Microorganisms 2020, 8, 355, 1-16 (2020).
- P. Kusari, S. Kusari, M. Spiteller and O. Kayser, Endophytic fungi harbored in Cannabis sativa L.: diversity and potential as biocontrol agents against host plant-specific phytopathogens. Fungal Diversity 60, 137–151 (2013).
- K. McKernan, Y. Helbert, L. Kane, N. Houde, L. Zhang, S. McLaughlin, Whole genome sequencing of colonies derived from cannabis flowers & the impact of media selection on benchmarking total yeast & mold detection tools, https://f1000research.com/articles/10-624 (2021).
- K. McKernan, Y. Helbert, L. Kane, L. Zhang, N. Houde, A. Bennett, J. Silva, H. Ebling, and S. McLaughlin, Pathogenic Enterobacteriaceae require multiple culture temperatures for detection in Cannabis sativa L. OSF Preprints, https://osf.io/j3msk/, (2022)



1. There is also no universal primer set that will amplify DNA of all bacterial or fungal species. Why are we concerned with counting organisms like powdery mildew for safety and compliance testing when they have extremely specific host requirements for growth like a metabolizing host plant? What’s the risks to humans if they can’t be responsible for opportunistic infection in a human host?
2. Why are compounds responsible for inhibition in culture-based techniques a major focus but compounds that inhibit PCR get ignored completely? If a compound interferes with polymerase enzymes or primer binding and knocks a Cq value back to a later cycle is that not a major concern in counting organisms with qPCR as well?
3. What is the mechanism that liberates a plant endophyte for quantitation in a qPCR assay that is any different from the mechanism used to liberate organisms from a sample for plating? If you can’t get these organisms out from inside plant tissues to count in plating, what allows you to get them out from plant tissues to be lysed and analyzed via qPCR?
4. Why is a method like DRBC plating, suggested for spreader molds in the FDA BAM, constantly used as the comparator for qPCR vs plating when it is known to be used specifically for certain types of organisms? Why does the 5-fold reduction of growth of aspergillus on DRBC media carry more weight than the 13-fold difference in rDNA gene copies found in different isolates of the same species of aspergillus fumigatus? https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0201611 If variation is a huge concern why no discussion on the variation of PCR reaction efficiency and lysis efficiency across different species? If molds react differently on different plating media why do we assume they react identically with one lysis approach?
I would think that if qPCR methods are advantageous over culture based methods for quantifying total yeasts and molds we would see more of them obtaining AOAC approval or use in any of the other various microbial testing industries. Why should cannabis testing be the canary in the coal mine for new methods with existing solutions?
Edward,
Good questions.
1. All Fungi have ITS sequences. All Bacteria have 16S sequences. 98% of microbes don’t culture. This is the reason CDC and the FDA are moving toward Culture Independent Testing (https://www.cdc.gov/foodsafety/challenges/cidt.html). While it may be hard to find a single primer set to universally amplify them all, it’s far more comprehensive than just capturing 2% of them like culture. Muliplex testing improves this. Additionally, primer sequences can be used to verify that your primers capture 1,000 of organisms where you can possibly buy this many organisms from a biobank to verify your inclusion criteria for plating.
2. Powdery mildew is just one example of a mold that doesn’t culture. But, as the article cites, most don’t culture. OSHA/CDC has listed powdery mildew and botrytis listed as an allergen for cannabis trimmers (Grant et al).
3. We use a combination of Lysis buffers and enzymatic digestion. These steps are noticeably absent in plating methods. It is far easier to find conditions that universally lyse organisms than conditions that universally grow them all. Zymo Research has good literature on this topic.
4. The FDA BAM lists 6 different media for TYM all of which provide different CFU for a given sample. We agree DRBC is the wrong media as do the reviewers of our paper on this topic (https://f1000research.com/articles/10-624/v2)
5. This is a red herring argument. The ability of an organism to grow or not grow on a given carbon source is a binary event (0 or 1). The fact that many organisms have 10 to 20 ITS regions simply alters the sensitivity of the test to sub CFU sensitivity. There are not fungi with zero ITS regions. Culture has more variables than just grow or not grow. Those that do grow, grow differently at different temperatures. CFU counts on plates will vary from 24 hours to 144 hours. Carbon source alters the count. Background matrix loaded with antibiotics alters the count. The background matrix is PCR is purified so the matrix variance seen with diverse cannabis chemotypes isn’t in play. The variance in ITS regions is small in comparison to culture and never dips to zero.
6. Many molecular tests are through AOAC approval and most companies with plating technologies have recently acquired molecular technologies. You will notice plating was abandoned for the C19 pandemic because it simply doesn’t scale and is too variable. It does however serve as an important benchmarking tool for molecular methods. They do need to be calibrated against organisms that do culture. We just don’t believe in gold standards with 98% blind spots.